FuSiOn Tutorial
Query a perturbagen pair
-
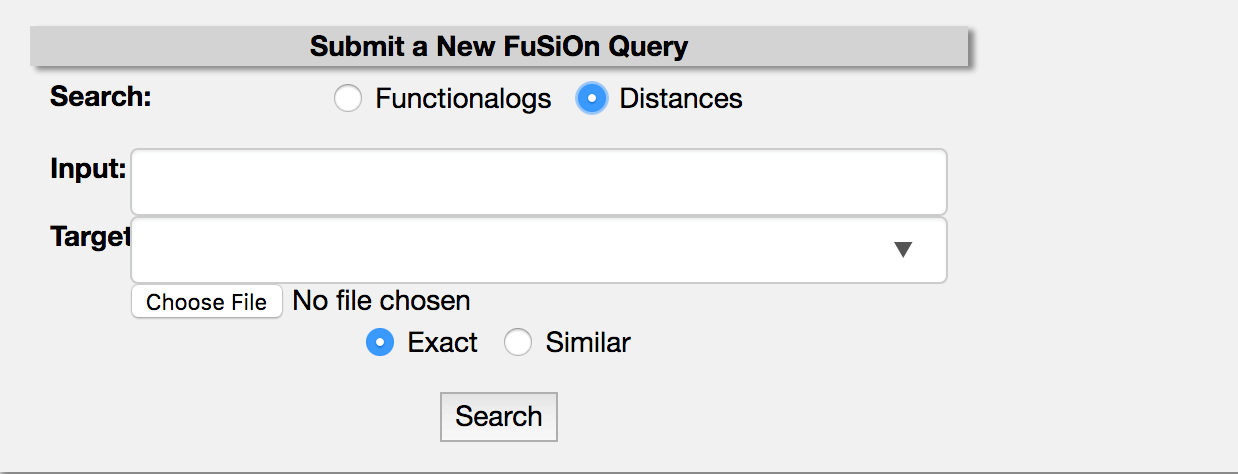
Select the button corresponding to ‘distances’ in the search field. Gene names will be auto-filled.
-
Enter a gene of interest into the ‘input’ box and search for distances to a query in the ‘target’ box. Distances, related statistical tests for the query pair, and a heatmap of the functional signature will be returned. Distances and FuSiOn signatures can be downloaded with the ‘Download Distances’ and ‘Download Expression’ buttons.
Find functionalogs to a gene query
-
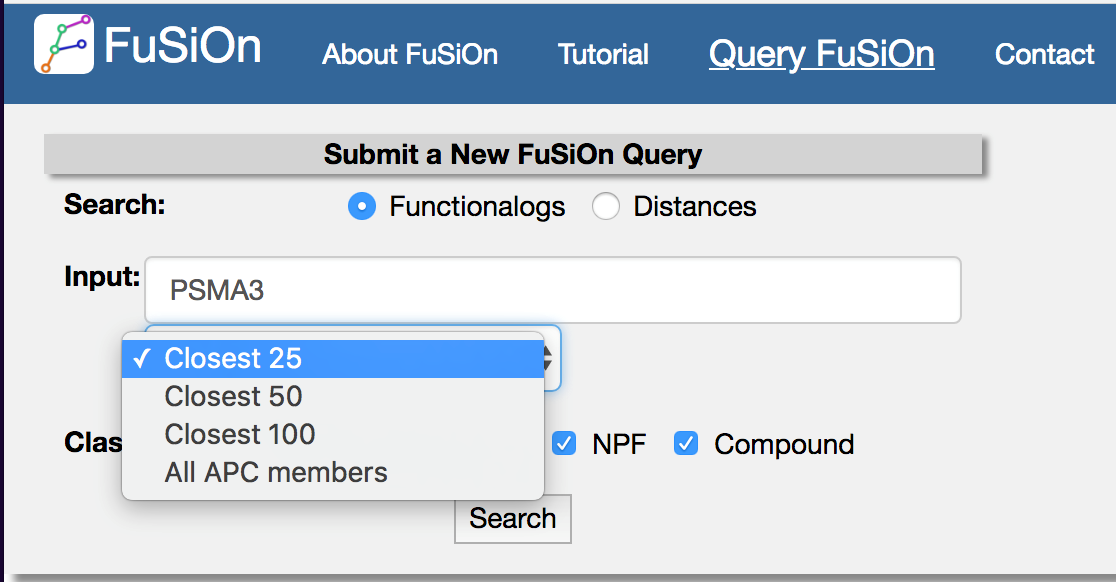
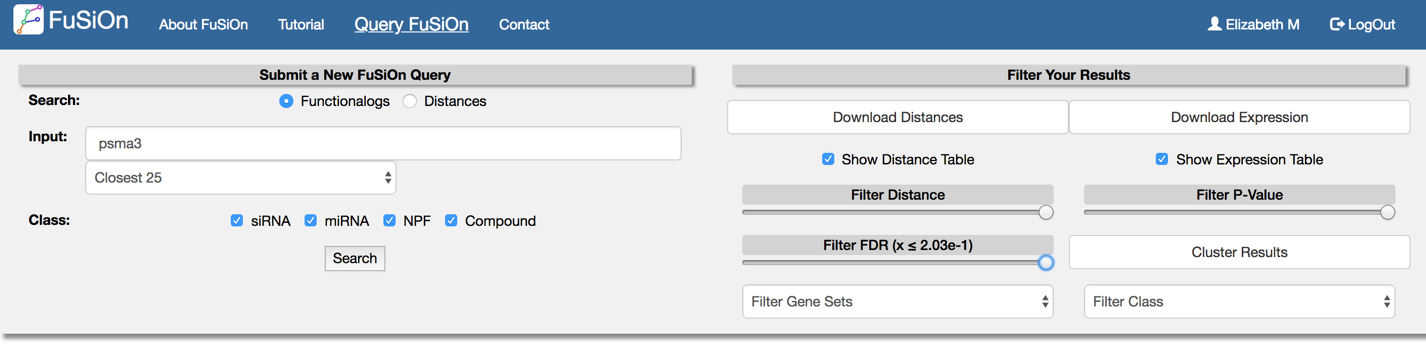
Enter a query gene, and select the number of closest functionalogs (by Euclidean distance) to return from the drop down menu.
-
Select one or more perturbagen datasets from the ‘class’ button to return functionalogs from the selected classes.
-
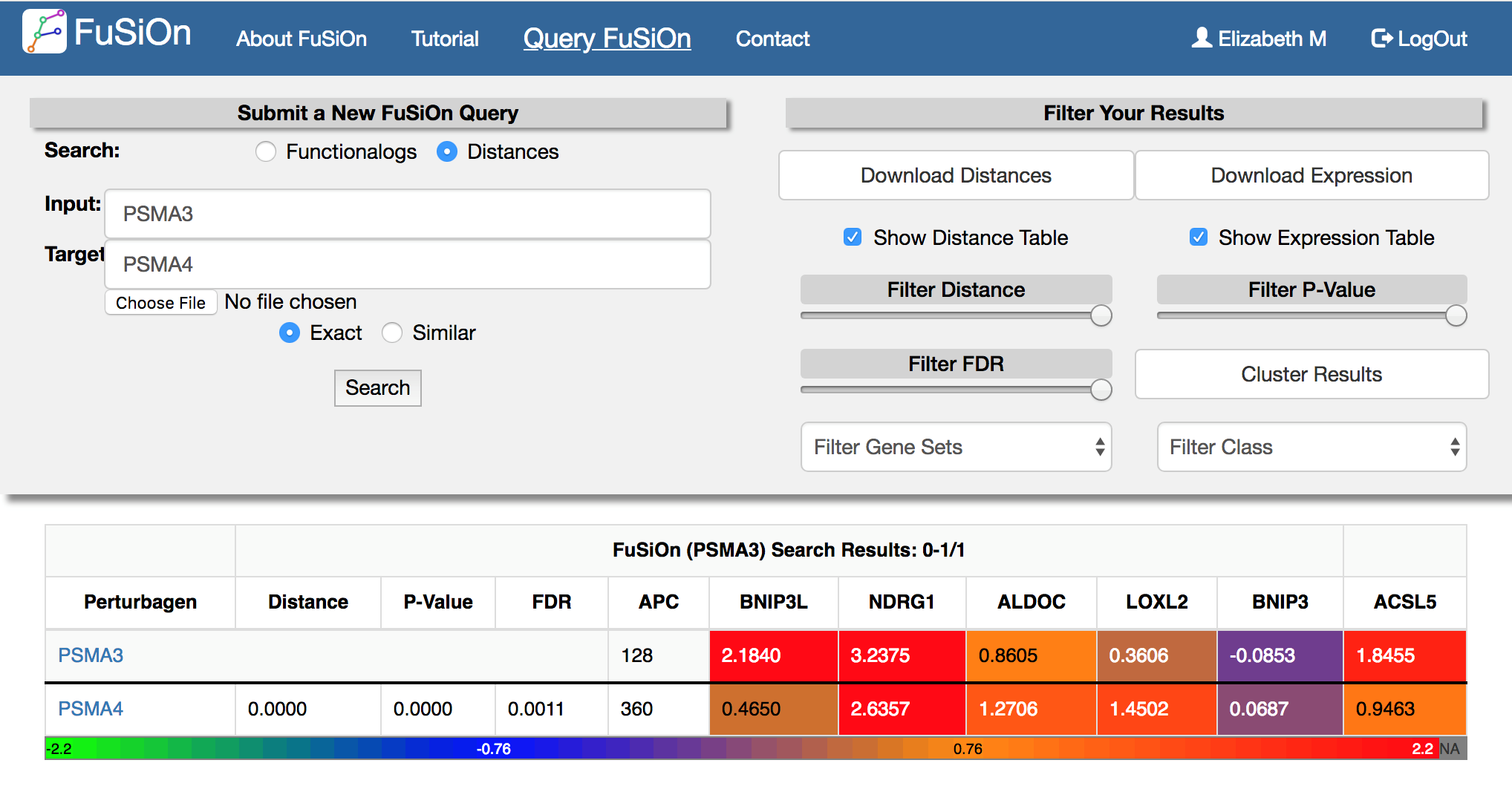
FuSiOn will return selected functionalogs, ranked by distances, together with associated statistics, clutering information (affinity propogation cluster number), and functional signature heatmaps. The query will be listed as the first line.
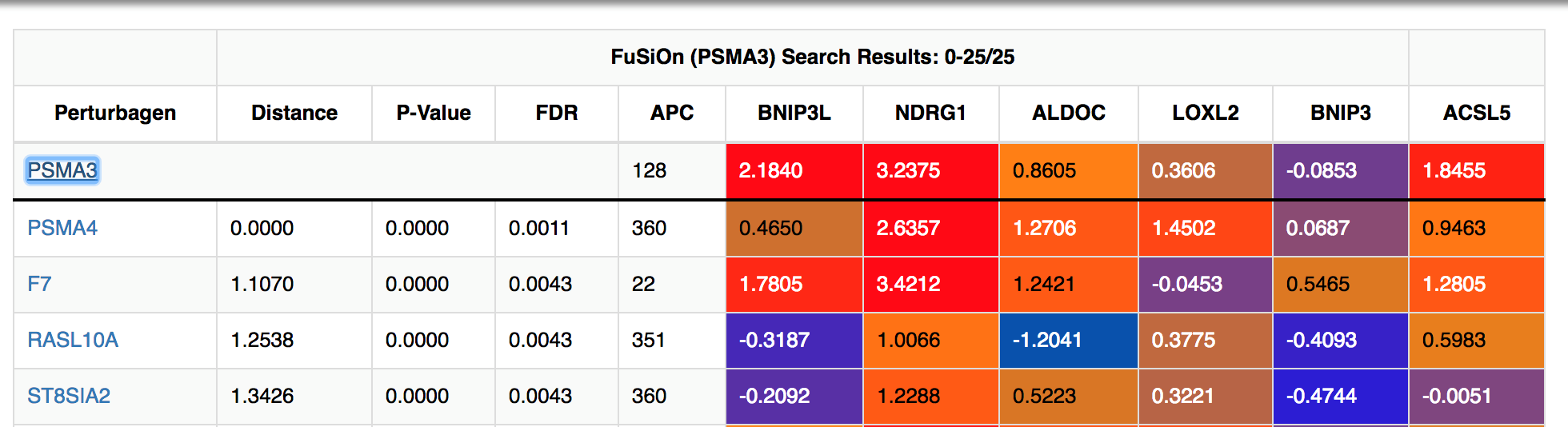
-
Toggle the ‘Filter Distance’, ‘Filter P-Value’, and ‘Filter FDR’ sliding bars to slow results less than user-defined cutoffs.
-
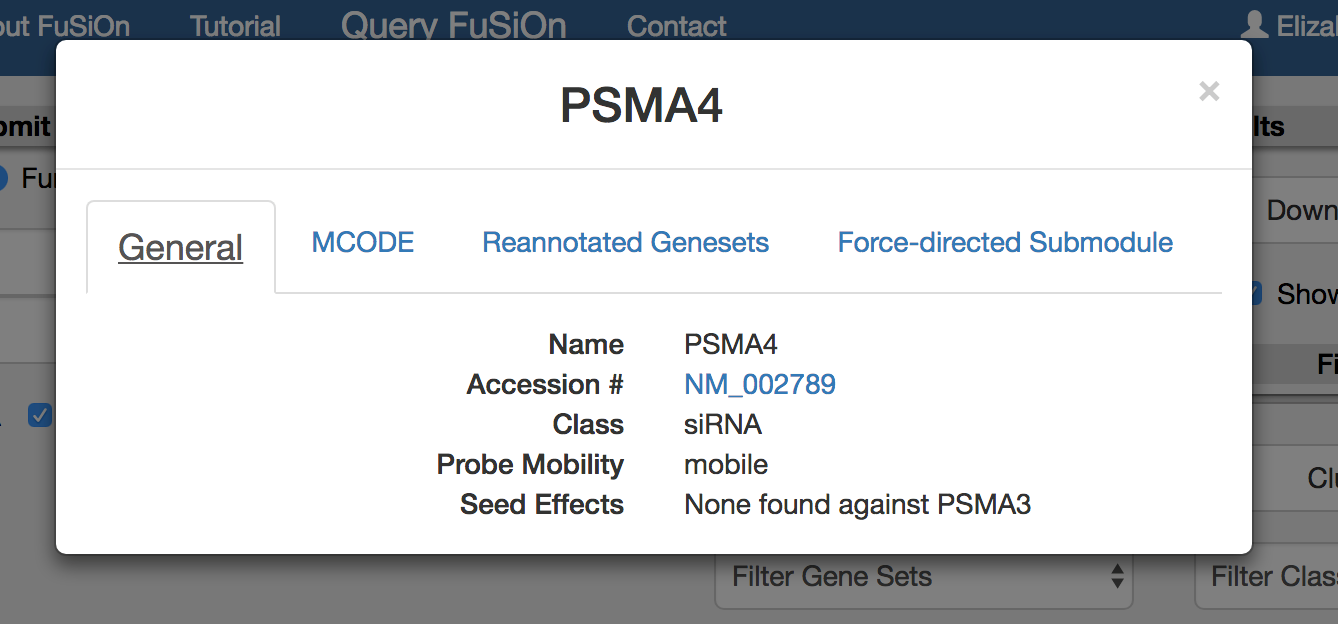
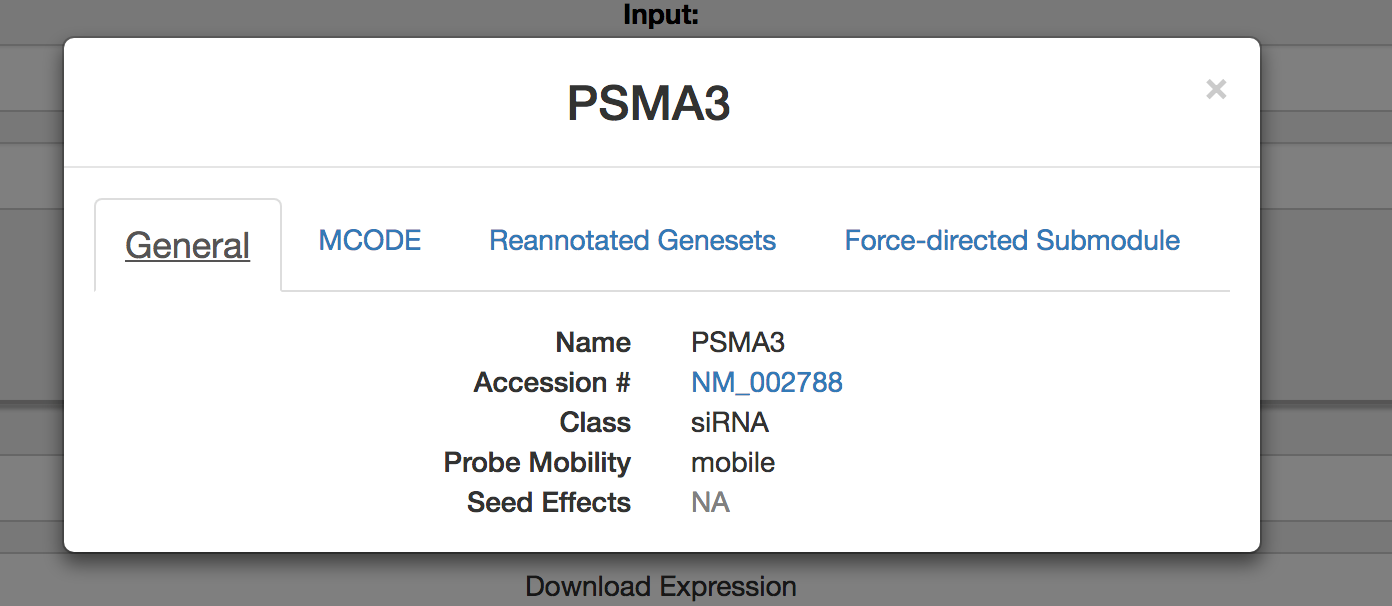
Select a gene to gain more information about the query, including information regarding reporter gene movability. A probe will be annotated as ‘immobile’ only if all 6 reporter genes do not significantly deviate from the mean reporter value for all tested perturbations. If the query and gene are from the siRNA dataset, information regarding clustering based on seed effect will be displayed.
-
Select the ‘cluster results’ button to display a 2-way hierarchical cluster for all genes returned in the window.
Query gene pathways discoverable in FuSiOn
-
Query a gene pair or functionalogs to a gene as described. Select on a gene by clicking on gene name for an information pop-up.
-
Click on the MCODE tab (if applicable) to discover protein-protein interactions enriched amongst the top 500 closest functionalogs to a query.
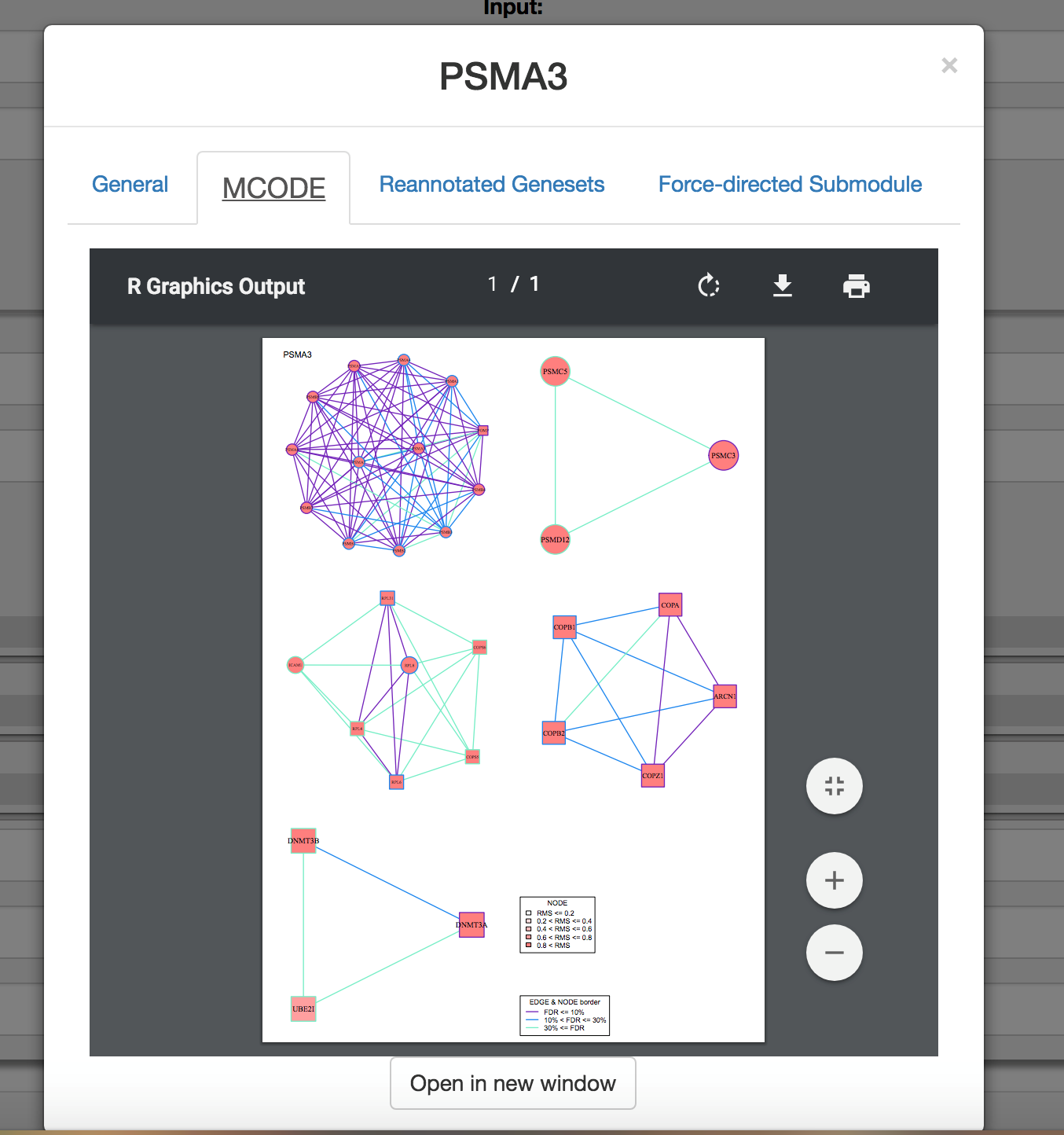
-
Click on the ‘Reannotated gene sets’ tab (if applicable) to discover if the query is present in a gene set determined to be discoverable within FuSiOn. All members within the pre-annotated gene sets listed have been determined to have FuSiOn distances that are significantly close..
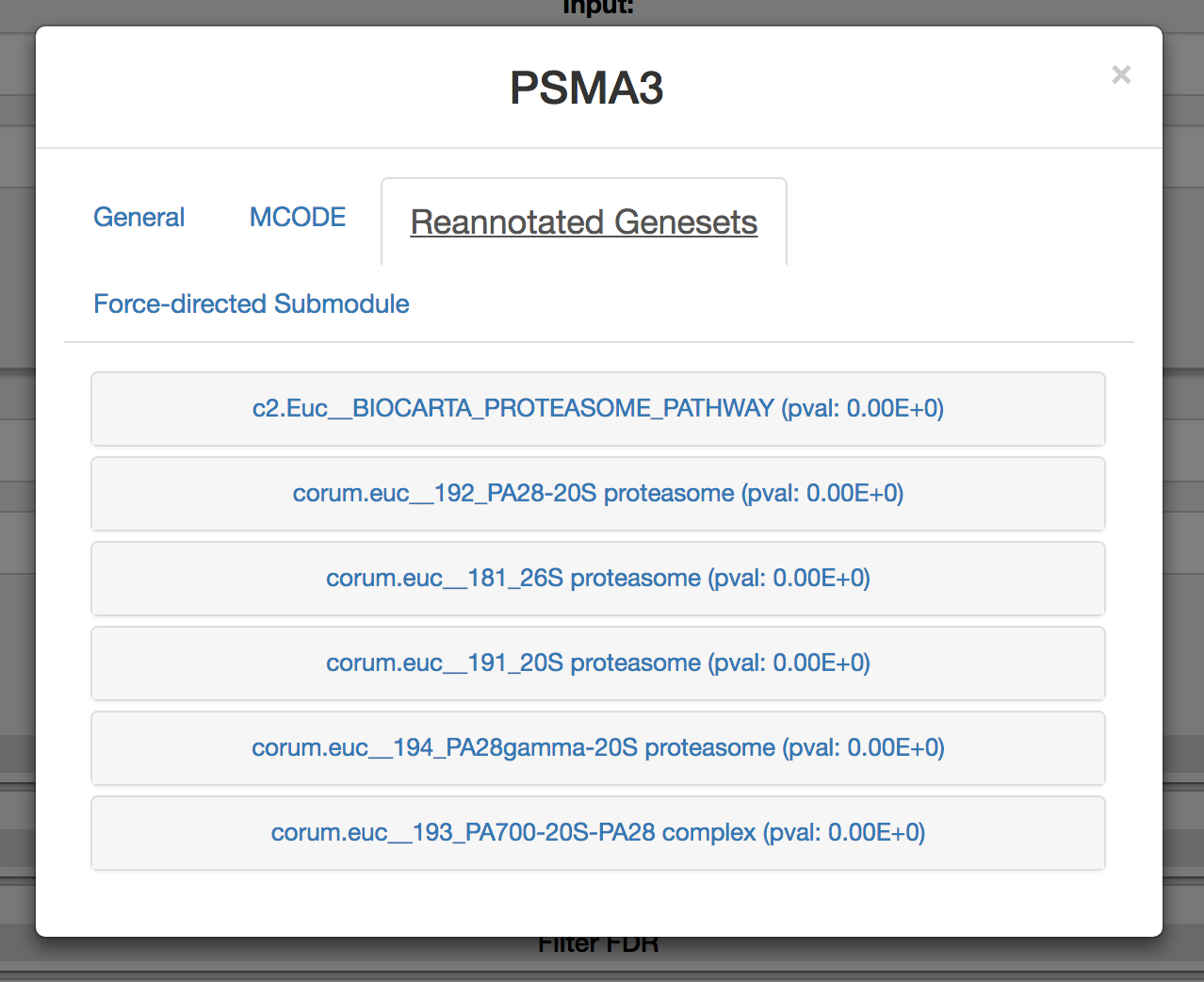
-
Click on a gene set to discover members. Members of the gene set are colored in red. Members colored in blue are genes outside the set with similar functional signatures..
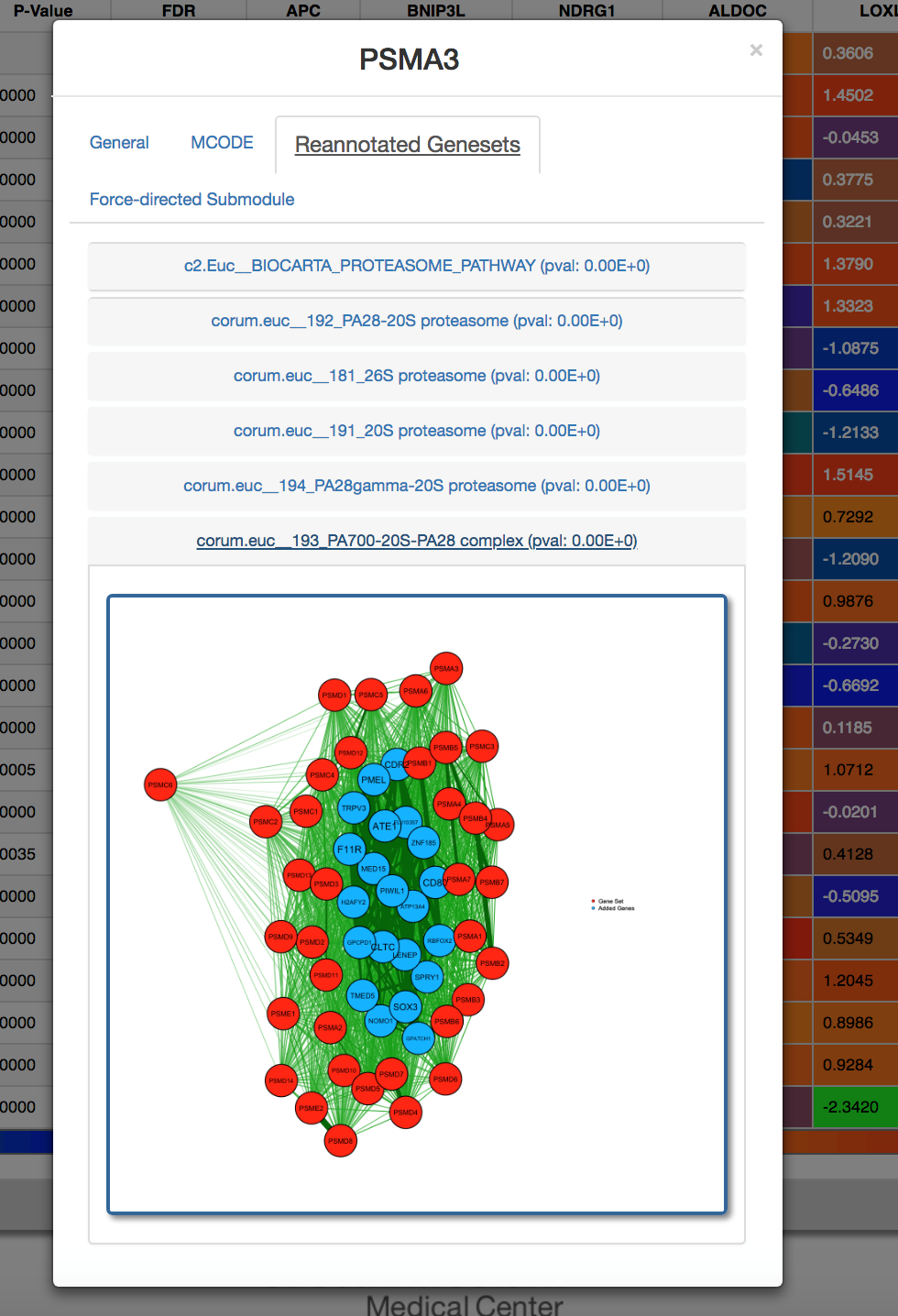
-
Select a gene set from the ‘Filter Gene Sets’ drop down menu on the main page to select genes only present in the chosen gene set.
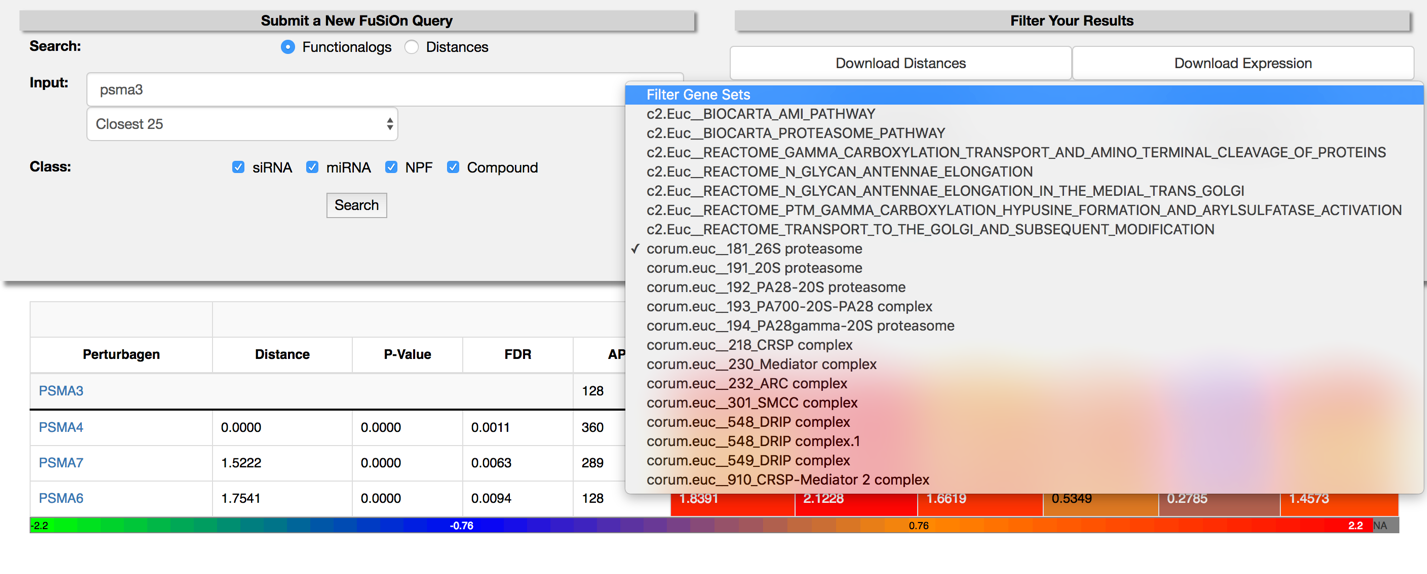
-
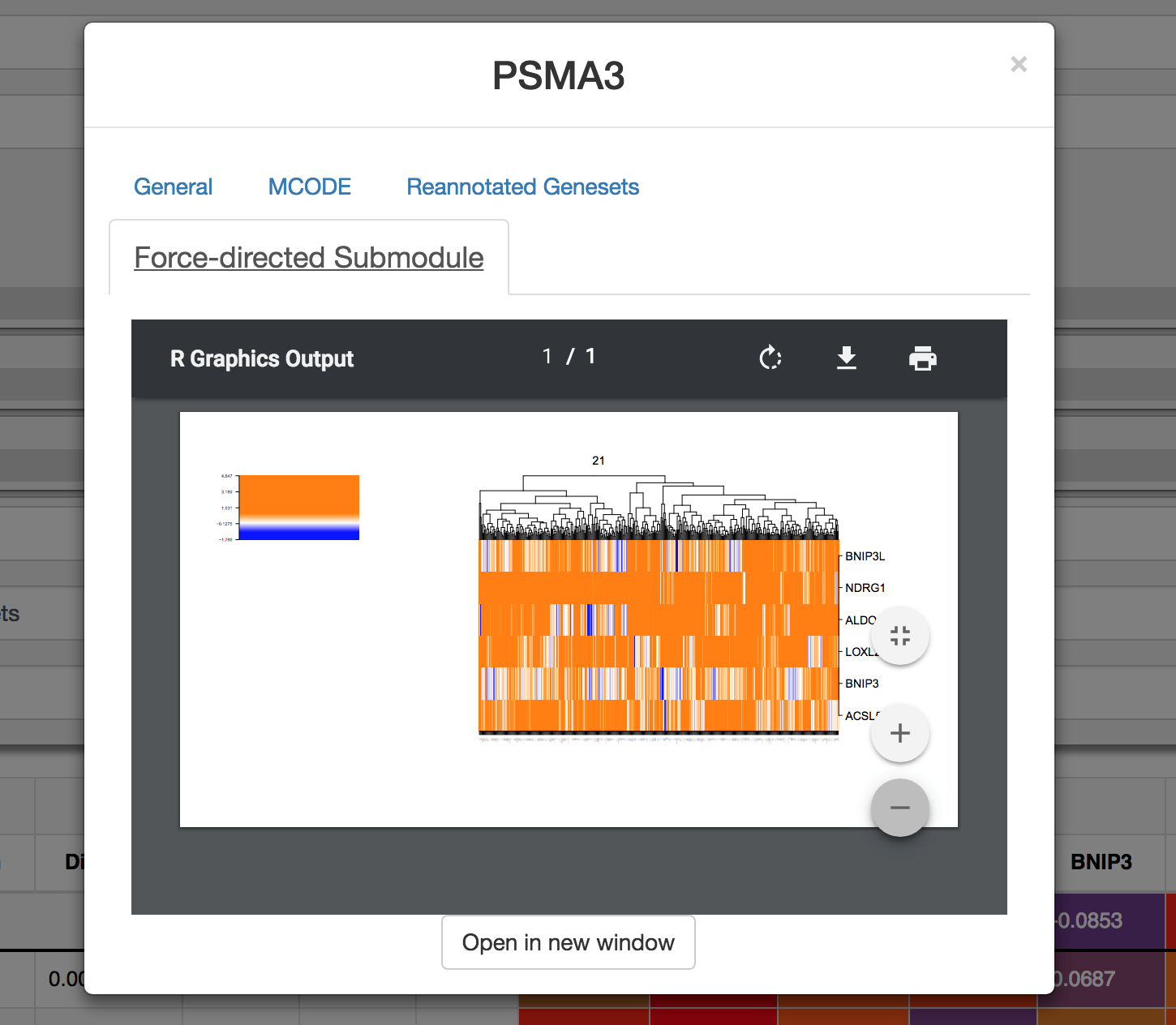
FuSiOn similarities were also used for de-novo gene network construction. Click on the ‘force directed submodule’ tab (if applicable) to see a submodule in which a gene was assigned. A representative heatmap of the FuSiOn signatures of all genes in the submodule will be displayed..